Hybrids
H. x nickeriensis Maas and de Rooij
Paulista - PE
Heliconia
BC- yellow-orange; OV- dark yellow
( H. marginata x H. psittacorum)
Pendulae distally, light yellow proximally, PD-
yellow, SE- dark yellow
H. psittacorum L.f x H. spathorcircinata Paulista - PE
uncertain
BC- yellow-Red; OV- yellow, PD-
Aristeguieta cv. Golden Torch
yellow with indistinct blackish green
Adrian
area distally, SE- dark yellow
H. psittacorum L.f x H. spathorcircinata Paulista - PE
uncertain
BC- yellow; OV- yellow, PD- yellow
cv. Golden Torch
with indistinct blackish green area
distally, SE- dark yellow
H. psittacorum L.f. x H. spathocircinata Paulista - PE
uncertain
BC- orange; OV- yellow, PD- yellow,
cv. Red Opal
SE- dark yellow with indistinct
blackish green area distally
Triploidd
H. psittacorum L.f.
Paulista - PE Stenochlamys BC- pink-green; OV- orange distally,
cv. Suriname Sassy
Stenochlamys yellow proximally, PD- orange to
cream, SE- orange with indistinct
blackish green area distally
H. psittacorum L.f. cv. Sassy
Paulista - PE Stenochlamys BC- pink-green; OV- orange distally,
Stenochlamys yellow proximally, PD- yellow
green, SE- orange with indistinct
blackish green area distally
Heliconia sp. (suposed to be H.
Maceió - AL Stenochlamys BC- pink-lilac; OV- green distally
psittacorum cv. Sassy)
Stenochlamys and yellow green proximally, PD-
yellow green, SE- orange with
indistinct blackish green area distally
Suposed triploid
H. psittacorum L.f . cv. Strawberries
Paulista - PE Stenochlamys BC- pink-yellow; OV- yellow to
and Cream
Stenochlamys cream, PD- cream, SE- pale yellow
with green spot on distally corner
H. psittacorum L.f. cv. Lady Di
Ubatuba - SP Stenochlamys BC- red; OV- yellow, PD- light yellow
Stenochlamys to cream, SE- light yellow with distally
dark green band and white tip
H. psittacorum L.f. cv. St. Vincent Red Ubatuba - SP Stenochlamys BC- red-orange; OV-orange distally, Stenochlamys orange to cream proximally, PD-orange, SE- orange with indistinct
blackish green area distally
H. psittacorum L.f. cv. Red Gold
Paulista - PE Stenochlamys BC- red-orange; OV- yellow, PD-
Stenochlamys yellow, SE- dark yellow with
indistinct blackish green area distally
a Identification based on Berry and Kress (1991) and Castro et al. (2007); b Based on Kress et al. (1993); c BC: bract color; OV: ovary; PD: pedicel; SE: sepals. d Ploidy (Costa et al., 2008).
Table 1. Genotypes, location, classification and description for 11 Heliconia psittacorum cultivars and interspecific hybrids of the UFRPE Heliconia Germplasm Collection used in this study
14
Agricultural Science
Molecular markers analyses occurred in the Plant Biotechnology Laboratory - UFRPE. The
optimization of the DNA extraction protocol was performed using fresh young leaves
samples of heliconia, harvested in the earliest stage of development and treated under three conditions: harvested, packed in a polystyrene box containing liquid nitrogen and taken to the Laboratory for immediate DNA extraction; harvested and frozen at -20°C for 1 day
before extraction; harvest and preserved in silica gel for 5 days before extraction.
In the DNA extraction, Doyle and Doyle (1990) protocol were used with modifications,
which was prepared at a 2x CTAB (hexadecyltrimethylammonium bromide) buffer solution.
It was added 700 microliter extraction buffer to 200 mg of macerated leaves in test tubes and taken to bath at 65°C. The tubes, after cooled at room temperature, were centrifuged and the supernatant transferred to new tubes. Supernatant was added to 700 microliter (L) CIA
(Chloroform-Isoamyl Alcohol) and then centrifuged was performed.
The supernatant was added to 700 microliter (L) CIA (Chloroform-Isoamyl Alcohol) and
then centrifuged. After this process, supernatant was added to 500 L of cold isopropanol and stored for 24 hours in a freezer at -20ºC. Subsequently, it was washed twice with 70%
ethanol and with 95% ethanol. The precipitate was dried at room temperature for 20
minutes and then resuspended with 300 L TE containing RNAse, incubated at 37°C for 30
minutes, then 5M NaCl and 300 L of cooled isopropanol were added, in which the DNA
was precipitated. Solution was incubated at 4°C throughout the night and the pellet
resuspended in 300 L TE. The DNA was quantified in 0.8% agarose gel.
Number of
Primers
Sequence
Basis
ITS 1
5’- TCCGTAGGTGAACCTGCGG -3’
19
ITS 2
5’-GCTGCGTTCTTCATCGATGC-3’
20
ITS 3
5’-GCATCGATGAAGAACGCAGC -3’
20
ITS 4
5’-TCCTCCGCTTATTGATATGC-3’
20
ITS 5
5´-GGAAGTAAAAGTCGTAACAAGG -3´
22
EF11 5-GTGGGGCATTTACCCCGCC-3'
19
EF22 5´-AGGAACCCTTACCGAGCTC-3´
19
trnL
5´-GGTTCAAGTCCCTCTATCCC -3´
20
trnF
5´-ATTTGAACTGGTGACACGAG-3´ 20
trnS
5´-TACCGAGGGTTCGAATC -3´
17
rps5’
5´-ATGTCCCGTTATCGAGGACCT -3´
21
rps3’
5’ –ATATTCTACAACTAACAACTC – 3’
21
Table 2. Sequence of primers used in amplification reactions in genotypes of the UFRPE
Heliconia Germplasm Collection used in this study
A set of 12 primers (Table 2) and 10 combinations of this primers were selected and tested for the ITS analysis: ITS1-ITS4; ITS5-ITS4; ITS1-ITS2; ITS5-ITS2; ITS3-ITS4 based on White et Genetic Diversity Analysis of Heliconia psittacorum Cultivars
and Interspecific Hybrids Using Nuclear and Chloroplast DNA Regions
15
al. (1990); and EF11-EF22; and for chloroplast genes analysis: rps3'-rps5' (Sanchez-Baracaldo, 2004) ; trnL-trnF (Sang et al. , 1997); trnS-trnF and trnS-trnL.
2.2 PCR amplification
The DNA amplification using PCR was performed to a final volume of 25 L containing 1
L template DNA, 0.3 L Taq-DNA polymerase (Invitrogen), 2.51 L Tris-HCl (pH 8, 0), and
0.75 L MgCl2, 2 L of each dNTPs, 1 L primer, 1 L oligonucleotide 1 and 2; and 15.45 L
milli-Q water to complete the reaction.
Amplifications were performed in a thermocycler MJ Reseach, Inc., PTC100 under the
following conditions: step 1 - following a denaturation step of 95°C for 3 minutes; step 2 –
94°C for 1 minute; step 3 – 58°C for 1 minute for annealing temperature; step 4 – 72°C for 1
min (repeat steps 2/3/4 for 29 cycles) followed by a final extension at 72°C for 10 minutes and 10°C for 24h. The PCR product visualization was performed in 1.5% agarose gel stained with SYBER Gold (Invitrogen), visualized under ultraviolet light and recorded on a digital Vilber Lourmat photographer.
2.3 Statistical analysis
Through the interpretation of gels, molecular data were tabulated as presence (1) or absence (0) of DNA fragments by primers for each genotype. Genetic similarities among genotypes
were determined based on the Jaccard (1908) coefficients. A dendrogram was then
constructed using the unweighted pair-group method of the arithmetic average (UPGMA)
based on the similarity matrix. The cluster analyses were conducted using the computer
program Gene (Cruz, 2006).
3. Results
The best condition for heliconia DNA extraction was using leaves in the earlyest stage of development, harvested, packed in a polystyrene box containing liquid nitrogen and taken
to the Laboratory for immediate DNA extraction.
3.1 Primers selection
Primer combination ITS4-ITS3 resulted in most of the polymorphic band region, while for
the primer combination ITS4-ITS5 it was observed the least polymorphism. The primers
used amplified from 1 to 6 band regions, with clear polymorphism between the genotypes.
The amplifications of the nuclear region that includes the spacers ITS1-ITS2 and EF11-EF22
(Fig. 1) generated fragments of approximately 396 to 506 pb, which agrees with Baldwin et al. (1995), by claiming that ITS markers have numerous small sized copies, reaching up to 700 pb.
Chloroplast regions amplifications that used the primers tRNA of leucine and phenylalanine ( trnL-trnF) generated fragments of approximately 1636 pb (Fig. 2). For the spacers regions rps3' - rps5' as well as for the regions trnS-trnL and trnS-trnF, it was observed monomorphic and polymorphic band patterns for the evaluated cultivars and hybrids.
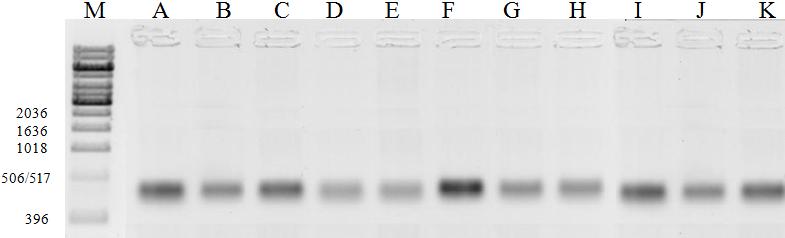
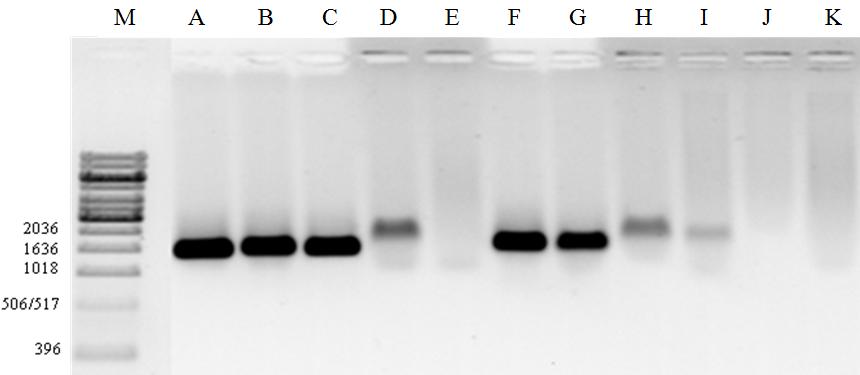
16
Agricultural Science
Fig. 1. Nuclear region amplifications that includes the spacers EF11-EF22. Cultivars and
interspecific hybrids: A- H. psittacorum cv. Sassy; B- H. psittacorum cv. Red Gold; C- H.
psittacorum x H. spathocircinata cv. Golden Torch Adrian; D- H. psittacorum cv. Suriname Sassy; E- H. psittacorum x H. spathocircinata cv. Red Opal; F- H. x nickeriensis; G- H.
psittacorum x H. spathocircinata cv. Golden Torch; H- Heliconia sp.; I- H. psittacorum cv. Lady Di; J- H. psittacorum cv. Strawberries e Cream; K- H. psittacorum cv. St. Vincent Red. (M = 1
kb DNA ladder).
Fig. 2. Chloroplast regions with primers trnL-trnF amplifications. Cultivars and interspecific hybrids: A- H. psittacorum cv. Sassy; B- H. psittacorum cv. Red Gold; C- H. psittacorum x H.
spathocircinata cv. Golden Torch Adrian; D- H. psittacorum cv. Suriname Sassy; E- H.
psittacorum x H. spathocircinata cv. Red Opal; F- H. x nickeriensis; G- H. psittacorum x H.
spathocircinata cv. Golden Torch; H- Heliconia sp.; I- H. psittacorum cv. Lady Di; J- H.
psittacorum cv. Strawberries e Cream; K- H. psittacorum cv. St. Vincent Red. (M = 1 kb DNA ladder).
3.2 Internal transcribed spacers
From the data generated by ITS markers and the analysis of the dendrogram (Fig. 3), it was observed the formation of two main groups (GI and GII) well sustained. The GI group, is
constituted by Heliconia sp., while, the other, more representative, GII, is subdivided into two other subgroups, SG A and SG B.
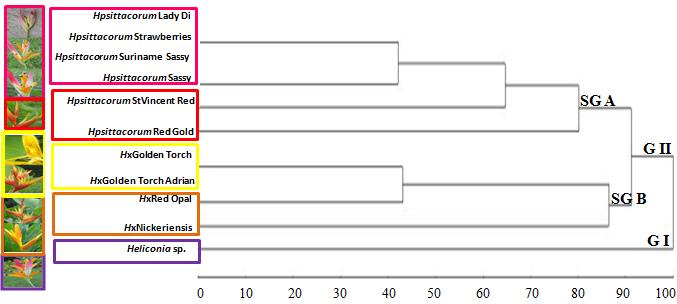
Genetic Diversity Analysis of Heliconia psittacorum Cultivars
and Interspecific Hybrids Using Nuclear and Chloroplast DNA Regions
17
Fig. 3. Cluster analysis on 11 genotypes of the UFRPE Heliconia Germplasm Collection,
used in this study, through ITS. G I= Group I; G II= Group II; SG A= Subgroup A; SG B=
Subgroup B.
The first group GI, consisting of Heliconia sp., that according to farmers, is identified as H.
psittacorum cv. Sassy and was more divergent from the other genotypes. The hypothesis that it is a new cultivar is supported by the fact that it was the only one that came from the state of Alagoas. It presents floral features intermediate between the triploid cultivars (Costa et al., 2008) of the subgroup SG A, bracts of pink and lilac, which presents individual characteristics such as, ovarian (OV) green distally and yellow green
proximally, pedicel (PD) yellow green and sepals (SE) orange with indistinct blackish
green area distally.
In the second group GII, subgroup SG A, formed by triploid cultivars of H. psittacorum: cvs.
Suriname Sassy and Sassy, that present bracts with pink and lilac, was also included
genotypes from the state of São Paulo, H. psittacorum cv. St Vincent Red and H. psittacorum cv. Lady Di.
The subgroup SG B was formed by the hybrid H. psittacorum x H. spathocircinata cvs. Golden Torch, Golden Torch Adrian and Red Opal, with bracts yellow and red. In this subgroup on
an external position, it was observed H. x nickeriensis, that is supposed to be an hybrid between H. psittacorum x H. marginata.
The hybrids showed low levels of similarity, around 12% of these comparisons reached
levels above 50% (Table 3), probably because they are the result from supposed crosses
between genetically distant parents or even the influences of epigenetic factors.
18
Agricultural Science
Table 3. Genetic similarity between 11 cultivars of H. psittacorum and interspecific hybrids of the UFRPE Heliconia Germplasm Collection used in this study
Genetic Diversity Analysis of Heliconia psittacorum Cultivars
and Interspecific Hybrids Using Nuclear and Chloroplast DNA Regions
19
4. Discussion
It was not possible to obtain DNA with acceptable quality from Heliconia using the conventional methodology, as mentioned by Kumar et al. (1998), in an earlier study with molecular markers in heliconia.
4.1 Primers selection
Band patterns variation may be related to high occurrence rate of base substitution and the great possibility of indels accumulation (events of inserts and/or deletions of nucleotides), moreover, these sequences are difficult to identify (Albert et al., 2002). The study with a great number of genotypes aims to explain the inheritance of the chloroplast, which may
vary according to the subgenus and be useful for genetic diversity studies of the group.
These primers ( trnL-F) have been successfully used in genetic diversity analysis of Orchidaceae (Kocyan et al., 2004) and Bromeliaceae groups (Sousa et al., 2007).
4.2 Internal transcribed spacers
In the absence of more precise evidence, it was decided to keep the genotype, here called Heliconia sp., as a specie not yet identified. It is assumed as a new cultivar of H. psittacorum cv. Sassy that occurred due to different geographic conditions. In fact, this finding requires further studies. Other molecular markers can be used to solve this issue, as did Kumar et al.
(1998), that using RAPD, found that two triploid cultivars, Iris and Petra were the same
genotype. Sheela et al. (2006) by using RAPD, found that cvs. St Vincent Red and Lady Di, were also grouped in the same subgroup. Thus, assuming that these genotypes formed a
subgroup brother of triploid cultivars H. psittacorum cv. Sassy and cv. Suriname Sassy, presenting 2n = 36 (Costa et al., 2008), leads to the assumption that cvs. St Vincent Red and Lady Di are supposed to be triploid, corroborating with the similar banding pattern among these four genotypes in primer combination ITS3-ITS4.
The group that gathered the hybrids H. psittacorum x H. spathocircinata cvs. Golden Torch, Golden Torch Adrian and Red Opal was expected, once the nrDNA has biparental
inheritance, and it is a nuclear molecular marker. H. x nickeriensis belongs to the Heliconia subgenus and Pendulae section (Kress et al., 1993), this subdivision is based on the consistency of vegetative structure, and staminodes and style shape, especially in the
pending heliconia. H. marginata, alleged parent, has pending inflorescence, and yet, differ from other hybrids that are crosses between H. psittacorum x H. spathocircinata and belongs to the Stenochlamys subgenus and Stenochlamys section (Kress et al., 1993). Using RAPD markers to study genetic variability and relationship between 124 genotypes of the genus Heliconia, Marouelli et al. (2010), managed to gather interspecific hybrids of H. psittacorum in the same clade.
The hybrids showed small similarity that can be explained by the coevolution hypothesis,
which considers the great genetic diversity of the genotype in the center of origin, once in northeast Brazil is frequently encountered native populations of H. psittacorum. Moreover, there is a wide variety of H. psittacorum hybrids described in literature, especially H.
spathorcircinata, confirming the potential of this specie to form hybrids (Berry and Kress, 1991).
20
Agricultural Science
The influence of epigenetic factors in the phenotype of an organism and therefore in
obtaining hybrids of Heliconia should be an issue to be raised. Characteristics of the transmissibility of an individual to other generations are not only linked to genes, the cell should be considered with its cytoplasm, mitochondria and genetic material carried in its structure, as well as the organism as a whole, and the complexity of the environment
(Pearson, 2006). Another factor to be considered is the cytosine methylation of the genetic material, also responsible for gene silencing, causing changes in the phenotype, and
according to most recent works can be passed to subsequent generations, thus causing
greate genetic diversity among individuals of the same specie.
Routinely, new Heliconia species have been described and others have been included as synonyms on each revision of the genus or subgenus; but, there is still controversy among authors. This situation suggests the need for a careful review of this group, since the visual botanical identification, may lead to imprecise denomination for the species that are being cultivated.
Although some diversity studies about the Heliconiaceae family have been undertaken in
recent years, its classification remains opened, therefore, new genetic markers for the group are required to elucidate these classification issues. The results revealed that there was no repetition of genetic material among the cultivars and interspecific hybrids of H. psittacorum evaluated, indicating the necessity to use other regions that could provide potentially
informative characters. In conclusion, the genetic diversity nuclear and chloroplast DNA
regions observed to study in Heliconia psittacorum cultivars and interspecific hybrids, are information promising to be taken in account as a first step towards genetic improvement.
5. Acknowledgements
The authors thank the National Council of Scientific and Technological Development
(CNPq) and the Coordination for the Improvement of Higher Education (CAPES) for the
scholarship of the first author, the BNB for the financial support, the Bem-Te-Vi Farm, the RECIFLORA association, researcher scientist Dr. Carlos E. F. de Castro
Campinas Agronomic Institute (IAC) and trainees of the UFRPE Floriculture Laboratory.
6. References
Albert, B., A.Jonhson, J.Lewis, M.Raff, K.Roberts and P.Walter (2002) Molecular Biology of The Cell, 4nd edn. Garland Publishing, New York, p.1661.
Baldwin, B.G., M.J.Sanderson, J.M.Porter, M.F.Wojcichowiski, C.S.Campbell and M.J.
Donoghue (1995) The ITS Region of Nuclear Ribosomal DNA: A Valuable Source of
Evidence on Angiosperm Phylogeny. Ann. Missouri Bot. Gard. 82: 247–277.
Berry, F. and W.J.Kress (1991) Heliconia: An Identification Guide. Smithsonian Institution Press, Washington and London, p.334.
Bruns, T.D., T.J.White and J.W.Taylor (1991) Fungal molecular systematics. Ann. Rev. Ecol.
Sys. 22: 525–564.
Câmara, P.E.A.S. (2008) Developmental, phyilogenetic, taxonomic study on the moss genus
Taxitelium Mitt. (Pylaisiadelphaceae). PhD Tesis, University of Missouri, St. Louis.
Castro, C.E.F., C.Gonçalves and A.May (2007) Atualização da nomenclatura de espécies do gênero Heliconia (Heliconiaceae). R. Bras. Hortic. Ornam. Campinas, Brazil. 13: 38–62.
Genetic Diversity Analysis of Heliconia psittacorum Cultivars
and Interspecific Hybrids Using Nuclear and Chloroplast DNA Regions
21
Castro, C.E.F. and T.T.Graziano (1997) Espécies do Gênero Heliconia (Heliconiaceae). R. Bras.
Hortic. Ornam. Campinas, Brazil. 3: 15–28.
Costa, A.S., B.S.F.Leite, V.Loges, E.C.S. Bernardes and A.C. Brasileiro-Vidal (2008) Padrão de distribuição de bandas CMA3 e localização de sítios de DNAr 5S e 45S na análise de
acessos de Heliconia (Heliconiaceae). In: Proceeding of 54d Congress on Genetic, Salvador, Brazil.
Criley, R.A. and T.K.Broschat (1992) Heliconia: botany and horticulturae of new floral crop.
Hortic. Review, New York. 14: 1–55.
Cruz, C.D. (2006) Programa Genes: análise multivariada e simulação. Viçosa, UFV, p.175.
Donselman, H. and T.K.Broschat (1986) Production and postharvest culture of Heliconia psittacorum flowers in south Florida. FL. Lauderdale, USA, pp.272–273.
Doyle, J.J. and J.L.Doyle (1990) Isolation of plant DNA from fresh tissue. Focus. 12: 13–15.
ITEP (2008) http://www.itep.br/lamepe.ASP. ( in portuguese).
Jaccard, P. (1908) Nouvelles recherché sur la distribution florale. Bull. Soc. Vaud. Sci. Nat.
44: 223–270.
Jatoi, S.A., A.Kikuchi, M.Mimura, S.S.Yi and K.N.Watanabe (2008) Relationship of Zingiber species, and genetic variability assessment in ginger ( Zingiber officinale) accessions from ex-situ genebank, on-farm and rural markets. Breed. Sci. 58: 261–270.
Johansen, L.B. (2005) Phylogeny of Orchidantha (Lowiaceae) and the Zingiberales based on
six DNA regions. Syst. Botany. 30:106–117.
Kladmook M., S.Chidchenchey and V.Keeratinijakal (2010) Assessment of genetic diversity
in cassumunar ginger ( Zingiber cassumunar Roxb.) in Thailand using AFLP markers.
Breed. Sci. 60: 412–418.
Kocyan, Y.L., P.K.Qiu, E.Endress and A.Conti1 (2004) A phylogenetic analysis of






